BioByte 138: Understanding Codon Translation with EnCodon, LabOS AI-XR Co-Scientist Assists in Wet Lab, Pearl Outperforms AF3, and the Brain's Predictive Immune Response to Infectious Visual Cues
Deep Dives
Explore related topics with these Wikipedia articles, rewritten for enjoyable reading:
-
Codon usage bias
13 min read
The article discusses EnCodon, a foundation model for understanding codon translation and the 'contextual grammar of codon usage.' Understanding codon usage bias - why organisms prefer certain codons over synonymous alternatives - is fundamental to grasping why this model matters for mRNA therapeutics and vaccine development.
-
Root mean square deviation of atomic positions
10 min read
The article mentions Pearl achieving improvements on 'RMSD-based benchmarks' and discusses 'sub-angstrom accuracy' thresholds. RMSD is the standard metric for comparing predicted protein-ligand structures to experimental ones, and understanding this metric is essential for evaluating drug discovery model performance.
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
Stars by Wassily Kandinsky, 1938
What we read
Blogs
Introducing Pearl: The Next Generation Foundation Model for Drug Discovery [Genesis Research Team, Genesis Molecular AI, October 2025]
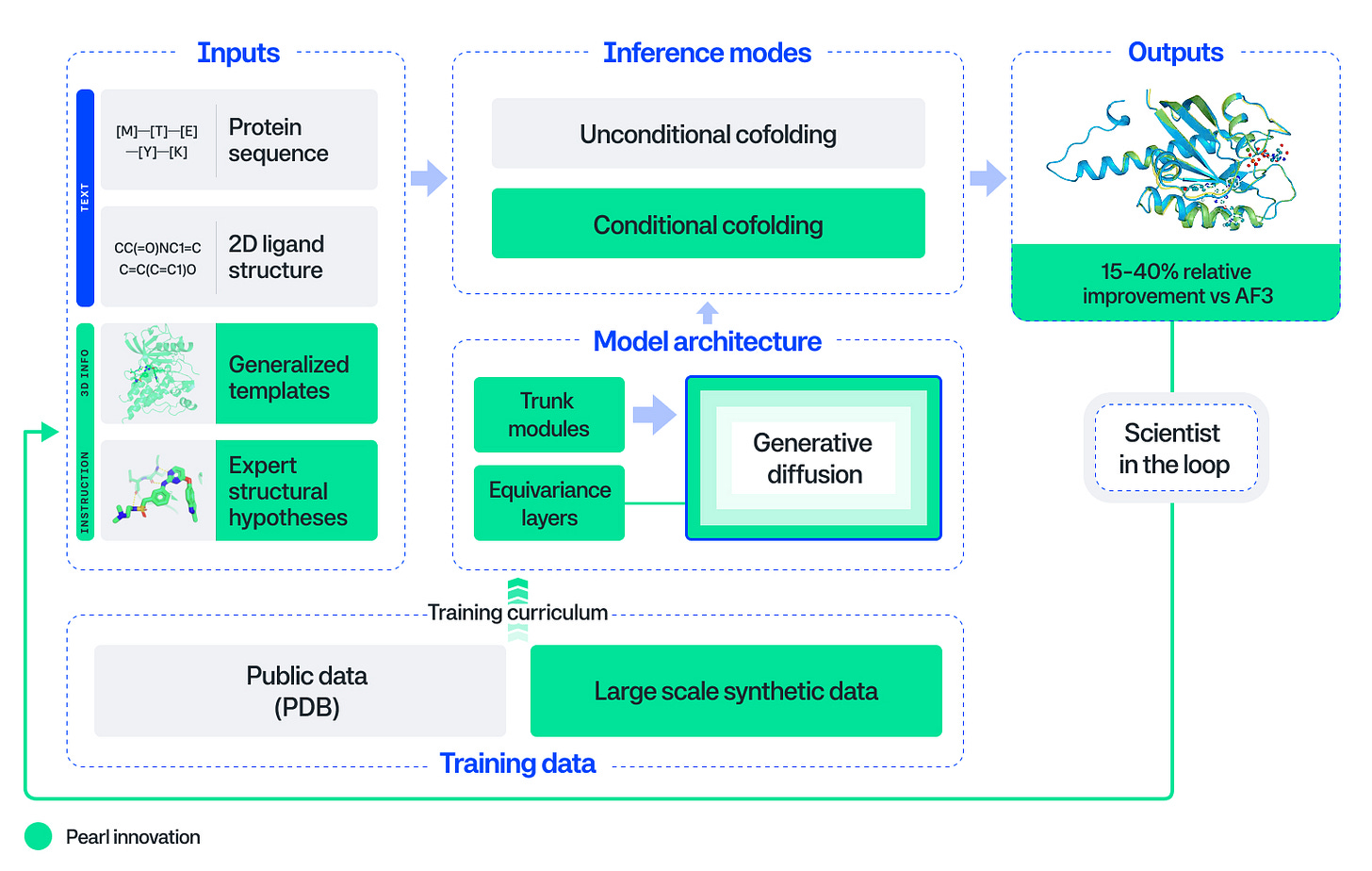
Accurately predicting how drugs bind to protein targets has been the holy grail of computational drug discovery. Genesis takes a significant step toward that goal: with Pearl, a protein-ligand structure prediction model, they surpass AlphaFold3 – achieving 14-15% relative improvements on standard RMSD-based benchmarks (PoseBusters and Runs N’ Poses) and substantially larger gains (3.6x at sub-angstrom accuracy) on proprietary, real-world drug targets. Beyond the headline numbers, Pearl’s explicit design for deployment is noteworthy: the model trains on large-scale synthetic complexes generated via physics simulations to overcome data scarcity, uses an SO(3)-equivariant diffusion architecture that enforces 3D rotational symmetry and geometric consistency in its predictions, and provides ‘scientist-in-the-loop’ control through templating and conditional inference modes enabling chemists to steer predictions with auxiliary structural information.
The technical choices reflect pragmatic engineering for drug discovery workflows rather than pure benchmark optimization. Training on synthetic physics-generated data is presented as a viable strategy – the team demonstrates that model performance scales monotonically with synthetic dataset size, suggesting a potential concrete path to improvement beyond the limited corpus of experimental structures. The conditional co-folding interfaces and templating system address a common failure mode of generative structure models: a lack of actionable control. Pearl allows chemists to leverage known binding pocket information or related crystal structures to generate and sample physically plausible poses they can iterate on – critical for lead optimization where sub-angstrom accuracy correlates with potency prediction. Importantly, the team demonstrates that standard <2Å accuracy thresholds are insufficient: qualitative analysis reveals that many poses below this threshold contain critical issues ranging from ring flips to missed interactions, rendering them spurious models for medicinal chemists.
The work comes with measurable caveats crucial to adoption in drug discovery. Reliance on proprietary and physics-generated training data raises questions about out-of-distribution robustness and whether synthetic priors fully capture binding thermodynamics – the associated paper itself catalogs common failure modes and emphasizes that high-accuracy thresholds are necessary for discovery utility. Moreover, independent validation will offer more decisive information on the model: blind prospective tests on withheld industrial targets, comparative analyses in
...This excerpt is provided for preview purposes. Full article content is available on the original publication.