BioByte 143: Predicting Structure of Large Protein Complexes, Restoring Naive T-cell Production, Large Scale Reading/Writing of Neuronal Activity, and Valuing Drug Programs with Tripartite Models
Deep Dives
Explore related topics with these Wikipedia articles, rewritten for enjoyable reading:
-
Thymus
12 min read
The article discusses thymic involution and its role in T cell production decline with age. Understanding the thymus's structure, function, and developmental biology provides essential context for the mRNA-LNP therapeutic approach described.
-
Icosahedral symmetry
14 min read
The Cosmohedra model exploits icosahedral symmetry to assemble viral capsid structures like dArc2. This geometric concept is fundamental to understanding why many viral proteins and large biological complexes share this specific symmetry pattern.
-
Lipid-based nanoparticle
12 min read
The Friedrich et al. study uses mRNA-LNP delivery to reprogram liver cells. Lipid nanoparticles are the key enabling technology for mRNA therapeutics, including COVID-19 vaccines, and understanding their mechanism illuminates this immunological research.
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
A Note To Our Readers: This is our last post for 2025 as we will be taking a short break from posting for the holidays. We’ll be back to our regular posting the first full week of January. Thank you from all of us for your support this year and stay tuned for more coming soon, Decoders! Happy holidays! ☃️
What we read
Papers
Scalable prediction of symmetric protein complex structures [Yu et al., bioRxiv, November 2025]
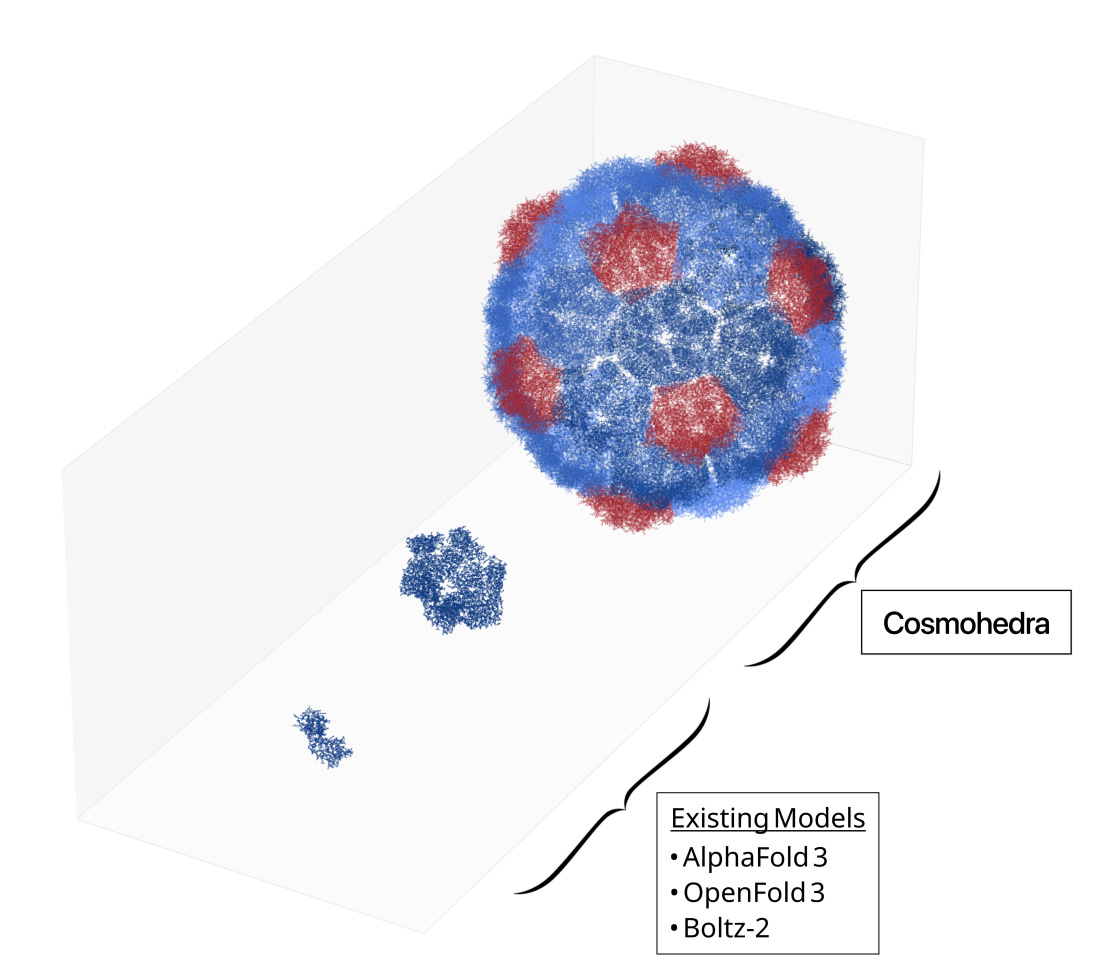
Why it matters: Researchers from the startup EndoFold built Cosmohedra, a physics-based model that assembles predicted structures of large, symmetric proteins at a fraction of the time previously required. This model unlocks biological inquiry and drug discovery into this class of previously expensive-to-model proteins - including building new drugs that disrupt viral complexes.
AlphaFold has been a breakthrough in protein structure prediction and downstream drug discovery, but it has its constraints. Chief among these is the large memory requirement: the transformers underpinning AlphaFold2/3 require memory that quadratically scales with sequence length (O(L2)), meaning that large proteins and protein complexes do not fit within a single AlphaFold prediction window without significant loss of accuracy or feasibility.
To tackle the problem of structure prediction for large proteins and complexes, the team at EndoFold take advantage of a convenient pattern in biology: protein symmetry. Many large complexes are just symmetric assemblies of smaller monomeric proteins! Take, for example, the Drosophila dArc2 capsid, a retrovirus-like capsid protein co-opted for neuronal cell-to-cell mRNA-based communication. The whole virus-like protein is an assembly of 240 identical Arc2 proteins, each 193 residues large. (Sidenote: Why does nature do this? One explanation is that it is genomically cheaper to encode a single monomer of 579 nucleotides, rather than explicitly encoding a full complex totaling 138,960 nucleotides.)
While AlphaFold can’t model the full complex, it generally can model the monomers of these large, symmetric proteins well! EndoFold builds on this foundation by constructing a physics-based assembly model Cosmohedra that uses true and predicted monomer structures and assembles them into symmetric complexes based on their symmetry class. To build the structure of the dArc2 capsid, for example, they assemble the predicted
...This excerpt is provided for preview purposes. Full article content is available on the original publication.