BioByte 140: Predicting Anti-Cancer Mechanisms of Action with DeepTarget, MOG-DFM Optimizes Therapeutic Peptide Design, and a New Means of Suppressing Inflammation in Autoimmune Diseases
Deep Dives
Explore related topics with these Wikipedia articles, rewritten for enjoyable reading:
-
Pareto efficiency
10 min read
The article discusses MOG-DFM's ability to produce 'Pareto-tuned candidate sets' and optimize across 'predicted Pareto fronts' - understanding Pareto efficiency and Pareto frontiers is essential for grasping how multi-objective optimization works in therapeutic peptide design
-
CRISPR gene editing
14 min read
DeepTarget's core methodology relies on comparing drug responses to genome-wide CRISPR-Cas9 knockout experiments to predict mechanisms of action - understanding how CRISPR knockouts work provides crucial context for the computational approach
-
Mechanism of action
10 min read
The article centers on predicting drug mechanisms of action in cancer treatment, discussing how many successful drugs have unknown MOAs - this Wikipedia article provides the pharmacological foundation for understanding why MOA prediction matters
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
Peter van der Doort, The Alchemist’s Laboratory (1595) [from Heinrich Khunrath, Amphitheatrum sapientiae aeternae]
A Note To Our Readers: Following this post, we will be taking a short break from posting, but will be back to regular posting the first week of December. We appreciate your support and stay tuned for more coming soon, Decoders!
What we read
Blogs
Designer Spotlight: Navigating the multi-property maze for therapeutic peptide design [Adaptyv Bio, Adaptyv Bio, November 2025 (based on original paper by Chen et al., arXiv, May 2025)]
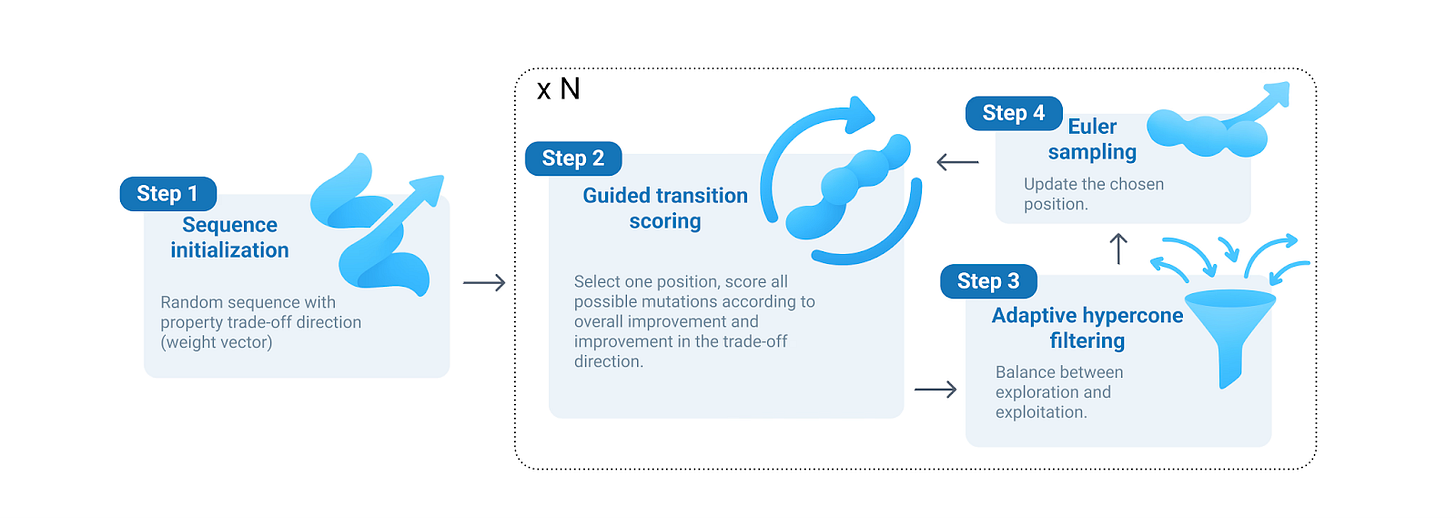
Biomolecular design is a multi-objective puzzle – therapeutic peptides face tradeoffs on properties such as affinity, stability, solubility, and safety. Navigating to the frontiers of feasible designs is a challenging task: properties are interdependent and sequence-phenotype relationships are non-linear and epistatic, forming a complex, high-dimensional landscape to traverse. Many generative models optimize for a single score and re-train for each desired objective, yielding a slow, brittle exploration of the design space. MOG-DFM (Multi-Objective Guided Discrete Flow Matching) offers a single pre-trained DFM generator that, at sampling time, can steer token-level proposals toward any user-specified set of objectives. This unlocks the ability to produce diverse, Pareto-tuned candidate sets without retraining for every tradeoff – a compelling capability for teams exploring competing design constraints.
The core development is a lightweight, local guidance rule: for each candidate token change, the algorithm computes a hybrid rank (relative candidate ordering on surrogate property predictors) and directional improvement score (expected improvement on the target tradeoff), measuring how a local edit shifts the multi-objective vector. These scores re-weight the DFM token transition rates so the continuous-time Markov chain sampler preferentially proposes beneficial edits. To avoid per-token ‘myopia’ or conflicting edits across multiple properties, the authors introduce an adaptive hypercone: an angular filter that requires proposed moves to lie within a tunable cone around a target objective direction, preserving coordination across tokens while allowing exploration.
The hypercone facilitates coordinated improvements across multiple properties and surpasses classical multi-objective optimization baselines on predicted Pareto fronts (hemolysis, half-life, solubility, binding, non-fouling). In silico, MOG-DFM shifts predicted trade-offs substantially (e.g. lower predicted hemolysis and higher solubility & half-life, while retaining affinity). Adaptyv provides wet-lab validation: of the 24 designed 10-mer peptides that were
...This excerpt is provided for preview purposes. Full article content is available on the original publication.