BioByte 129: strategies to drug intrinsically disordered proteins , in silico prediction of RNA-LNP efficacy, and minibinder design to address multidrug-resistant bacterial infections
Welcome to Decoding Bio’s BioByte: each week our writing collective highlight notable news—from the latest scientific papers to the latest funding rounds—and everything in between. All in one place.
What we read
Blogs
Hitting ‘undruggable’ disease targets [Baker Lab, Institute for Protein Design, July 2025]
Intrinsically disordered proteins and regions (IDPs / IDRs) are central to signaling and disease but have resisted conventional binder design since they lack a stable conformation. Through two complementary papers released by the Baker Lab, researchers reframe IDP plasticity as a design opportunity. The teams used generative protein design to induce and capture binding competent conformations, leading to high-affinity, high-specificity protein binders across in vitro and in vivo (cellular) experiments.
One study - published in Science - introduces the ‘logos’ strategy. This entails developing a library of scaffolds and pocket modules that can be recombined and tailored to match arbitrary disordered sequences. Targets are threaded through these templates, the best matches are refined with ML sequence design, and pockets are tuned with diffusion methods to achieve atomic complementarity. In a one-shot campaign, the authors reported binders for 39 of 43 diverse IDR targets (with typical Kd’s from ~100pM to 100nM, and testing ~22 designs per target) and demonstrated functional readouts, including a dynorphin binder that blocked opioid signaling in cultured human cells. The pipeline emphasizes modularity and combinatorial pocket assembly in order to provide a general solution to sequence-based recognition in IDPs.
The second study - published in Nature - focused on applying RFdiffusion to co-sample target and binder conformations, allowing dynamic molecular shifts during design to reveal favorable induced-fit complexes. Operating solely from sequence, the team generated nanomolar-affinity binders (~3-100 nM) for targets such as amylin, G3BP1, and prion core fragments. Functional efficacy is exhibited again - one such case involving amylin binders dissolving and disassembling amyloid fibrils and rerouting amylin to lysosomes.
Cumulatively, the papers offer a robust proof-of-principle for transforming disorder into a tractable engineering challenge. The performance of this approach under native cellular complexities (e.g. phase separation, post-translational modifications) and therapeutically-relevant constraints - such as developability testing - remains an interesting question for future investigation.
Papers
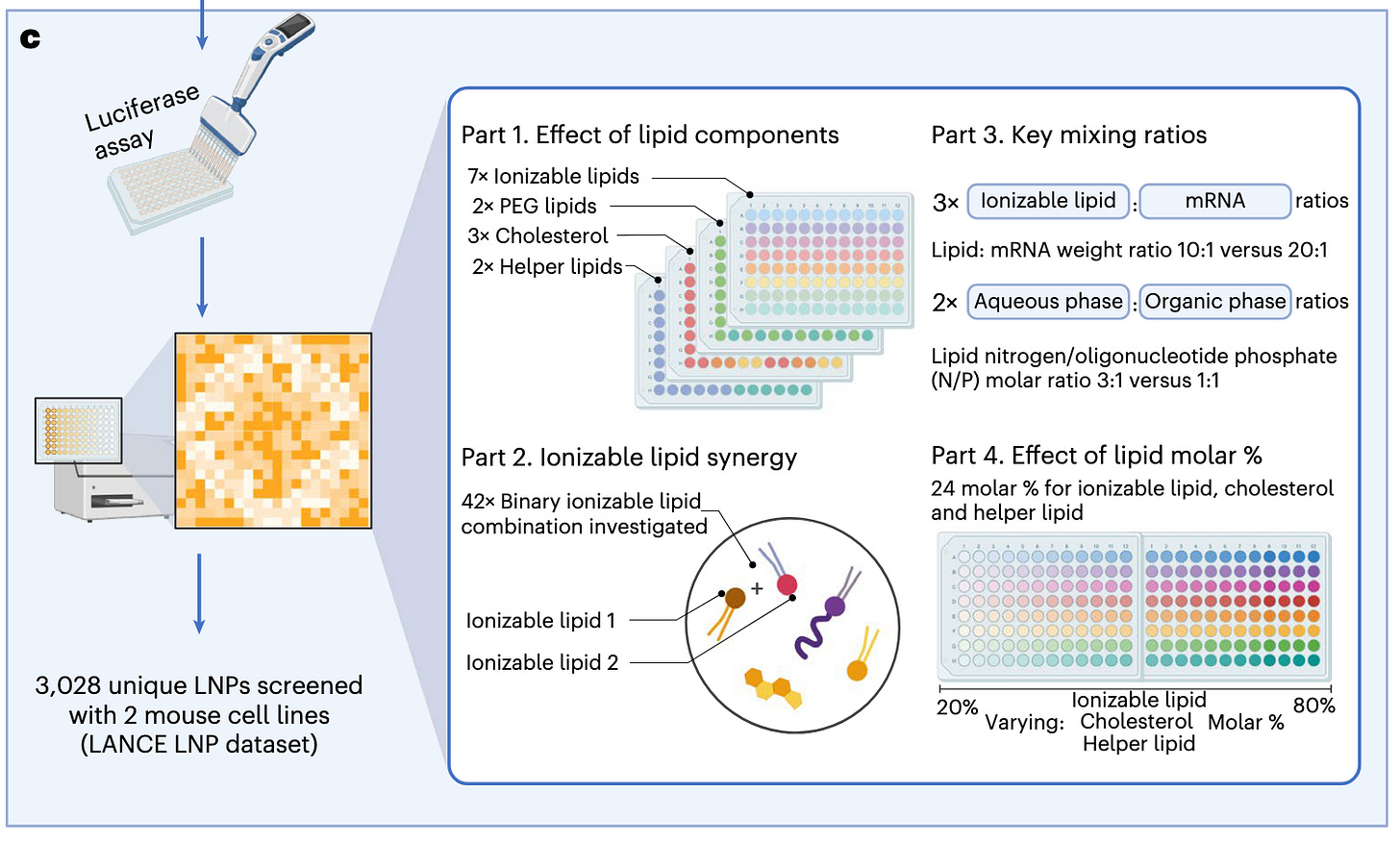
Designing lipid nanoparticles using a transformer-based neural network [Chan et al., Nature Nanotechnology, August 2025]
...Why it matters: RNA-LNPs are an increasingly important tool in the therapeutic arsenal. The effectiveness of this modality is determined, in part, by its lipid components and
This excerpt is provided for preview purposes. Full article content is available on the original publication.